DNA Nanotechnology Creates Dynamic Cytoskeleton for Artificial Cells
Dynamic Nucleic Systems Lab/UCLA
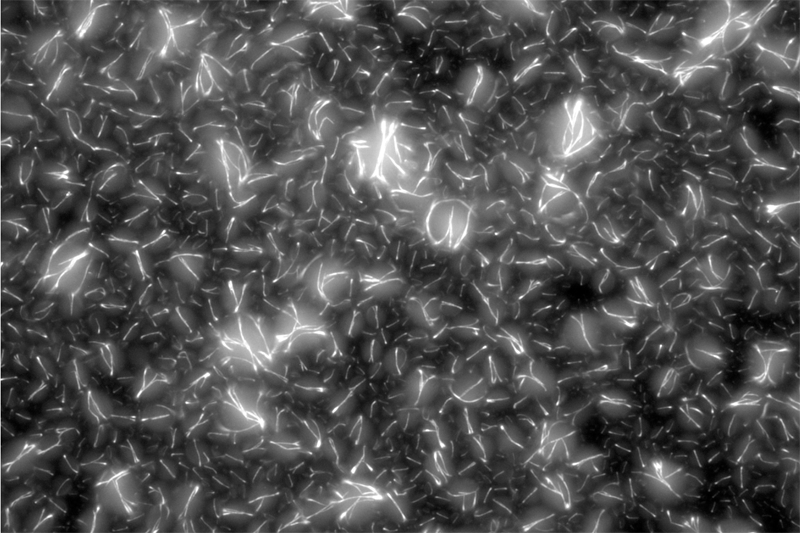
Fluorescence microscopy image of DNA nanotubes
Using DNA and RNA molecules, UCLA engineers have built a tiny artificial molecular scaffolding system that resembles the complex network of interlinking protein chains found in the cytoplasm of all living cells. The methodology could be customized to build scaffolds inside artificial cells capable of autonomous molecular functions.
Nature Communications recently published the group’s paper outlining the research.
Artificial, or synthetic, cells can be as simple as droplets of water in oil. They are used in biology to study how cells and their internal components work, and in medicine to develop and deliver drugs and therapies. They could be used to produce food or fuels, of even be harnessed for bio computers, which use biologically derived molecules to perform computations.

The majority of attempts at engineering molecular systems involve using natural biological scaffolds with complex protein molecules that were reconstituted inside an artificial cell. This approach poses many challenges as the numerous components of the cytoskeleton have high levels of interactions among themselves. The complex biological crosstalk is often not measurable, making it even more difficult to design the cells’ dynamic responses.
The UCLA-led research developed a more streamlined system that will enable artificial cells to adapt, partition and move.
“Our system is simpler to build as it has fewer molecule components compared to natural biological scaffolds,” said principal investigator Elisa Franco, an associate professor of mechanical and aerospace engineering at the UCLA Samueli School of Engineering. “This provides a unique toolkit that can be used to build a complex cytoskeleton that mimics life-like functions, and opens up new pathways for research across several fields that use artificial cells.”
The team first pre-programmed single strands of DNA molecules to connect with one another. This was done by determining the molecules’ sequences of constituent nucleotides — adenine binds with thymine and guanine binds with cytosine (or “A-T” and “G-C”). The respective single strands then came together to form interlocking tiles of DNA. These slightly curved tiles then locked in place either inside or outside of an artificial cell, forming long and flexible nanoscale tubes, or nanotubes, which constituted the artificial cytoskeleton.
In addition to showing how to build the self-assembling scaffolds, the researchers also demonstrated they could control when the system assembles and disassembles by incorporating RNA molecules in the process.
“Our research is a big step toward the bottom-up development of a nucleic acid-based cytoskeleton that mimics synthetic cells,” said Franco.
“Our research is a big step toward the bottom-up development of a nucleic acid-based cytoskeleton that mimics synthetic cells,” said Franco, who also teaches bioengineering and leads the Dynamic Nucleic Systems Lab at UCLA. “Due to the high modularity of these nucleic acid components, the scaffolds could include molecular sensors that allow detection of, and adaptability to, environmental and chemical changes.”
The advance will also enable researchers to customize an artificial cell’s internal organization systematically, with advanced mechanical properties that could facilitate artificial cell division.
The study’s co-first authors are Siddharth Agarwal and Melissa Klocke — engineering doctoral students at UCLA and UC Riverside, respectively. Passa Pungchai, a UCLA bioengineering undergraduate student, is also an author.
The U.S. Department of Energy supported the research. Some experiments were performed at the Advanced Light Microscopy and Spectroscopy Laboratory, which is part of the Leica Microsystems Center of Excellence in the California NanoSystems Institute (CNSI) at UCLA. The facility has been supported by the National Institutes of Health and the National Science Foundation.
